Bayesian Benefit Risk
benefit(name, fun, weight)
risk(name, fun, weight)
br(...)
mcda(...)Arguments
- name
a string indicating the name of the benefit or risk.
- fun
a utility function which maps a parameter value to a utility value.
- weight
the weight of the benefit/risk.
- ...
calls to
benefit(),risk(), andbr_group()to define the utility functions and treatment groups.
Value
A named list with posterior summaries of utility for each group and the raw posterior utility scores.
Details
The br() function allows the user to define an arbitrary number
of "benefits" and "risks". Each benefit/risk requires a utility
function (fun) and a weight. The utility function maps the benefit/risk
parameter to a utility score. The br_group() function supplies samples
from the posterior distribution for each benefit risk for a specific
group (e.g. treatment arm).
The br() function then calculates the posterior distribution of the
overall utility for each group. The overall utility is a weighted sum of
the utilities for each benefit/risk.
The mcda() function is the same as br(), but has extra checks to
ensure that the total weight of all benefits and risks is 1, and that the
utility functions produce values between 0 and 1 for all posterior
samples.
Examples
set.seed(1132)
ilogit <- function(x) 1 / (1 + exp(-x))
out <- mcda(
benefit("CV", function(x) ilogit(x), weight = .75),
risk("DVT", function(x) ilogit(- .5 * x), weight = .25),
br_group(
label = "PBO",
CV = rnorm(1e4, .1),
DVT = rnorm(1e4, .1)
),
br_group(
label = "TRT",
CV = rnorm(1e4, 2),
DVT = rnorm(1e4, 1)
)
)
out
#> # A tibble: 20,000 × 11
#> CV CV_weight CV_uti…¹ CV_sc…² DVT DVT_w…³ DVT_u…⁴ DVT_s…⁵ label iter
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <int>
#> 1 1.47 0.75 0.813 0.610 0.439 0.25 0.445 0.111 PBO 1
#> 2 0.239 0.75 0.559 0.420 -2.12 0.25 0.743 0.186 PBO 2
#> 3 0.969 0.75 0.725 0.544 0.974 0.25 0.381 0.0951 PBO 3
#> 4 0.655 0.75 0.658 0.494 0.0771 0.25 0.490 0.123 PBO 4
#> 5 1.51 0.75 0.819 0.615 -0.993 0.25 0.622 0.155 PBO 5
#> 6 -1.32 0.75 0.210 0.158 -0.874 0.25 0.607 0.152 PBO 6
#> 7 0.395 0.75 0.598 0.448 1.03 0.25 0.375 0.0936 PBO 7
#> 8 -1.43 0.75 0.193 0.145 -0.903 0.25 0.611 0.153 PBO 8
#> 9 0.180 0.75 0.545 0.409 -0.685 0.25 0.585 0.146 PBO 9
#> 10 -0.960 0.75 0.277 0.208 -1.50 0.25 0.679 0.170 PBO 10
#> # … with 19,990 more rows, 1 more variable: total <dbl>, and abbreviated
#> # variable names ¹CV_utility, ²CV_score, ³DVT_weight, ⁴DVT_utility,
#> # ⁵DVT_score
summary(out, probs = c(.025, .5, .975))
#> $summary
#> # A tibble: 2 × 5
#> label mean `2.50%` `50.00%` `97.50%`
#> <chr> <dbl> <dbl> <dbl> <dbl>
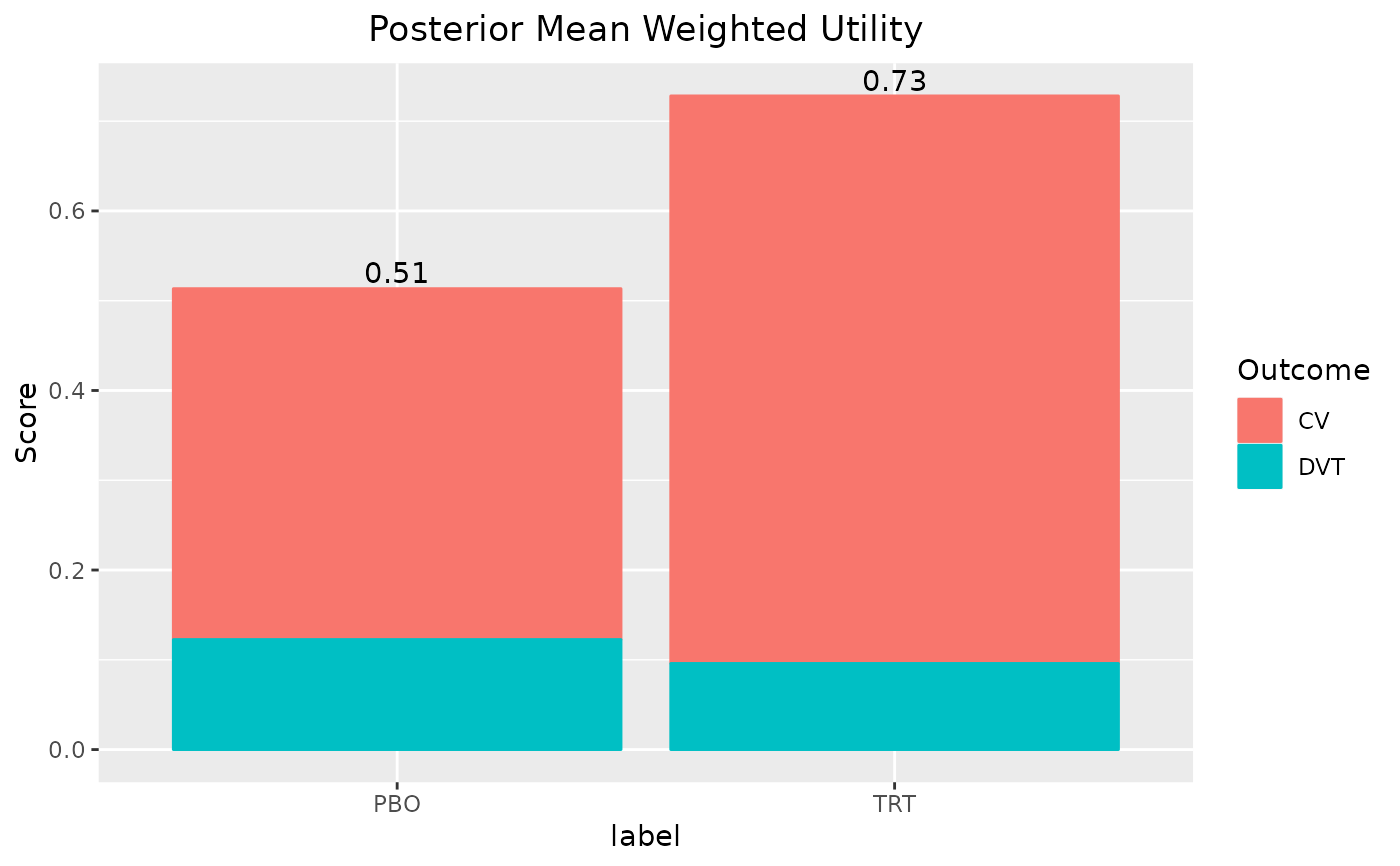
#> 1 PBO 0.513 0.221 0.518 0.794
#> 2 TRT 0.728 0.472 0.752 0.857
#>
#> $scores
#> # A tibble: 20,000 × 11
#> CV CV_weight CV_uti…¹ CV_sc…² DVT DVT_w…³ DVT_u…⁴ DVT_s…⁵ label iter
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <int>
#> 1 1.47 0.75 0.813 0.610 0.439 0.25 0.445 0.111 PBO 1
#> 2 0.239 0.75 0.559 0.420 -2.12 0.25 0.743 0.186 PBO 2
#> 3 0.969 0.75 0.725 0.544 0.974 0.25 0.381 0.0951 PBO 3
#> 4 0.655 0.75 0.658 0.494 0.0771 0.25 0.490 0.123 PBO 4
#> 5 1.51 0.75 0.819 0.615 -0.993 0.25 0.622 0.155 PBO 5
#> 6 -1.32 0.75 0.210 0.158 -0.874 0.25 0.607 0.152 PBO 6
#> 7 0.395 0.75 0.598 0.448 1.03 0.25 0.375 0.0936 PBO 7
#> 8 -1.43 0.75 0.193 0.145 -0.903 0.25 0.611 0.153 PBO 8
#> 9 0.180 0.75 0.545 0.409 -0.685 0.25 0.585 0.146 PBO 9
#> 10 -0.960 0.75 0.277 0.208 -1.50 0.25 0.679 0.170 PBO 10
#> # … with 19,990 more rows, 1 more variable: total <dbl>, and abbreviated
#> # variable names ¹CV_utility, ²CV_score, ³DVT_weight, ⁴DVT_utility,
#> # ⁵DVT_score
#>
summary(out, reference = "PBO")
#> $summary
#> # A tibble: 1 × 5
#> label mean `2.50%` `97.50%` reference
#> <chr> <dbl> <dbl> <dbl> <chr>
#> 1 TRT 0.215 -0.150 0.555 PBO
#>
#> $scores
#> # A tibble: 10,000 × 12
#> CV CV_weight CV_utility CV_sc…¹ DVT DVT_w…² DVT_u…³ DVT_s…⁴ label iter
#> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl> <chr> <int>
#> 1 1.01 0.75 0.733 0.550 0.381 0.25 0.453 0.113 TRT 1
#> 2 1.66 0.75 0.841 0.631 1.84 0.25 0.285 0.0713 TRT 2
#> 3 0.473 0.75 0.616 0.462 0.403 0.25 0.450 0.112 TRT 3
#> 4 2.59 0.75 0.930 0.698 0.0774 0.25 0.490 0.123 TRT 4
#> 5 2.39 0.75 0.916 0.687 2.47 0.25 0.225 0.0563 TRT 5
#> 6 2.43 0.75 0.919 0.689 1.08 0.25 0.368 0.0920 TRT 6
#> 7 1.58 0.75 0.828 0.621 2.02 0.25 0.267 0.0667 TRT 7
#> 8 3.10 0.75 0.957 0.718 1.04 0.25 0.373 0.0932 TRT 8
#> 9 3.48 0.75 0.970 0.728 0.644 0.25 0.420 0.105 TRT 9
#> 10 1.10 0.75 0.750 0.563 0.586 0.25 0.427 0.107 TRT 10
#> # … with 9,990 more rows, 2 more variables: total <dbl>, reference <chr>, and
#> # abbreviated variable names ¹CV_score, ²DVT_weight, ³DVT_utility, ⁴DVT_score
#>
plot(out)
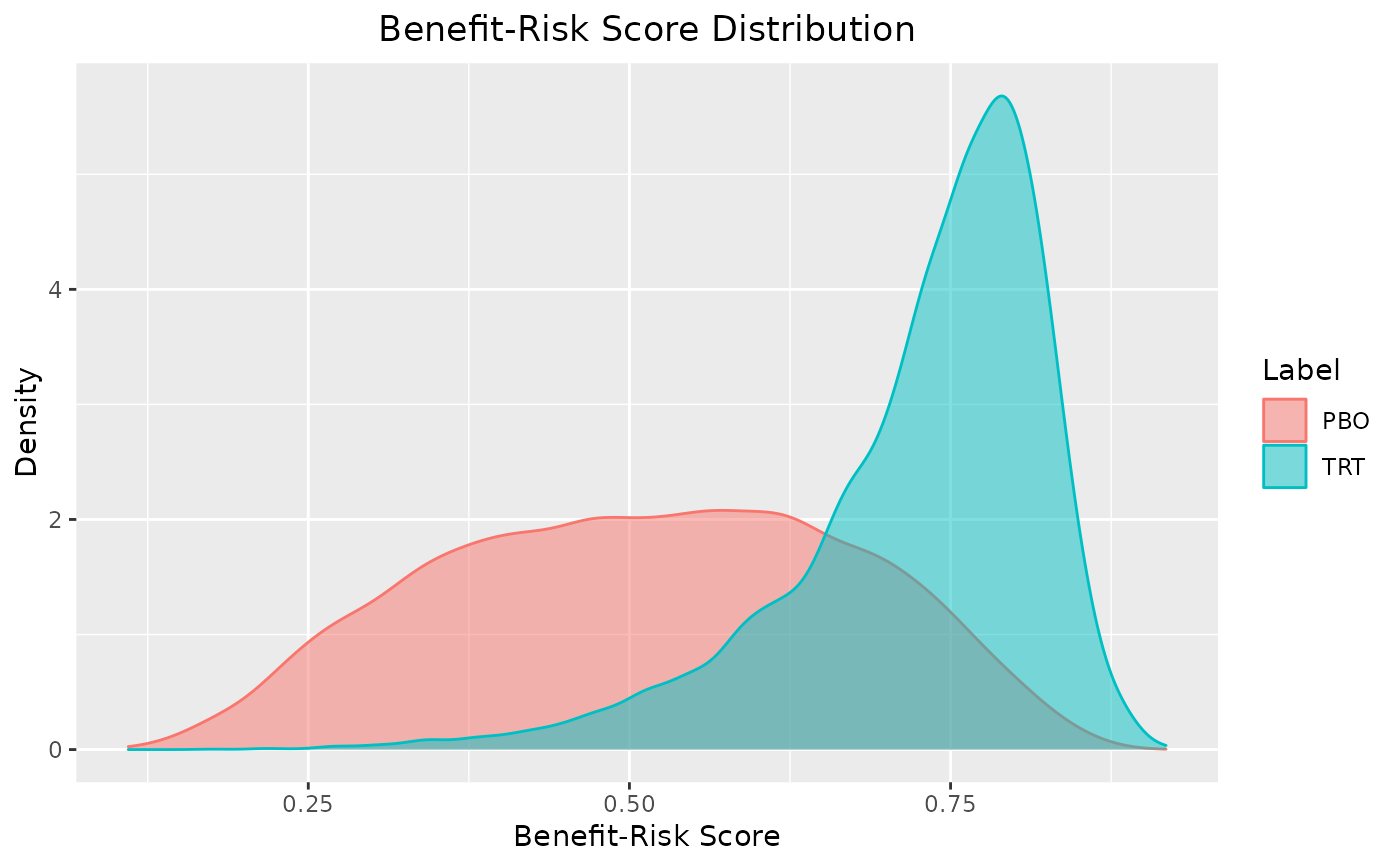 plot(out, reference = "PBO")
plot(out, reference = "PBO")
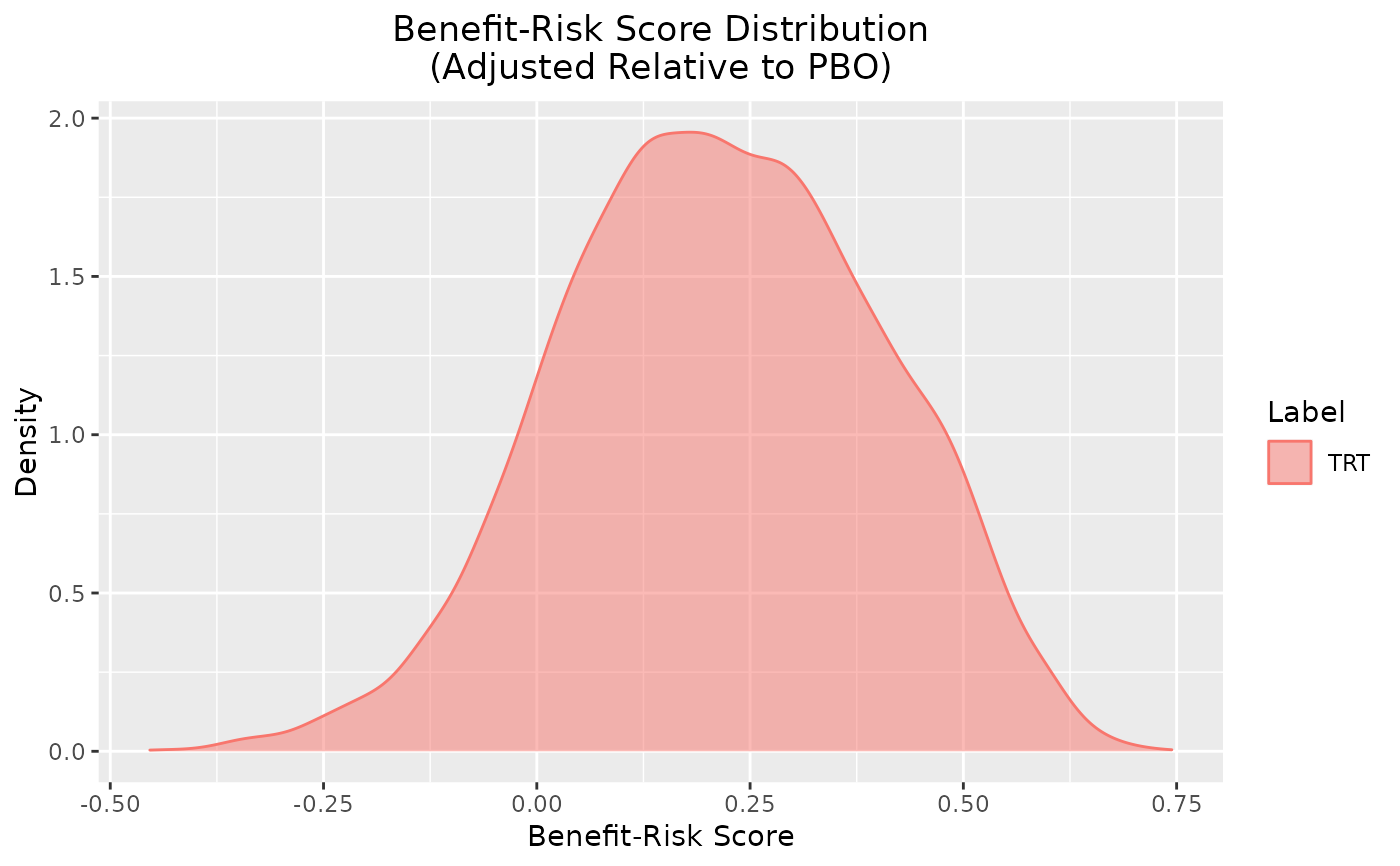 plot_utility(out)
plot_utility(out)
 plot_utility(out, reference = "PBO")
plot_utility(out, reference = "PBO")
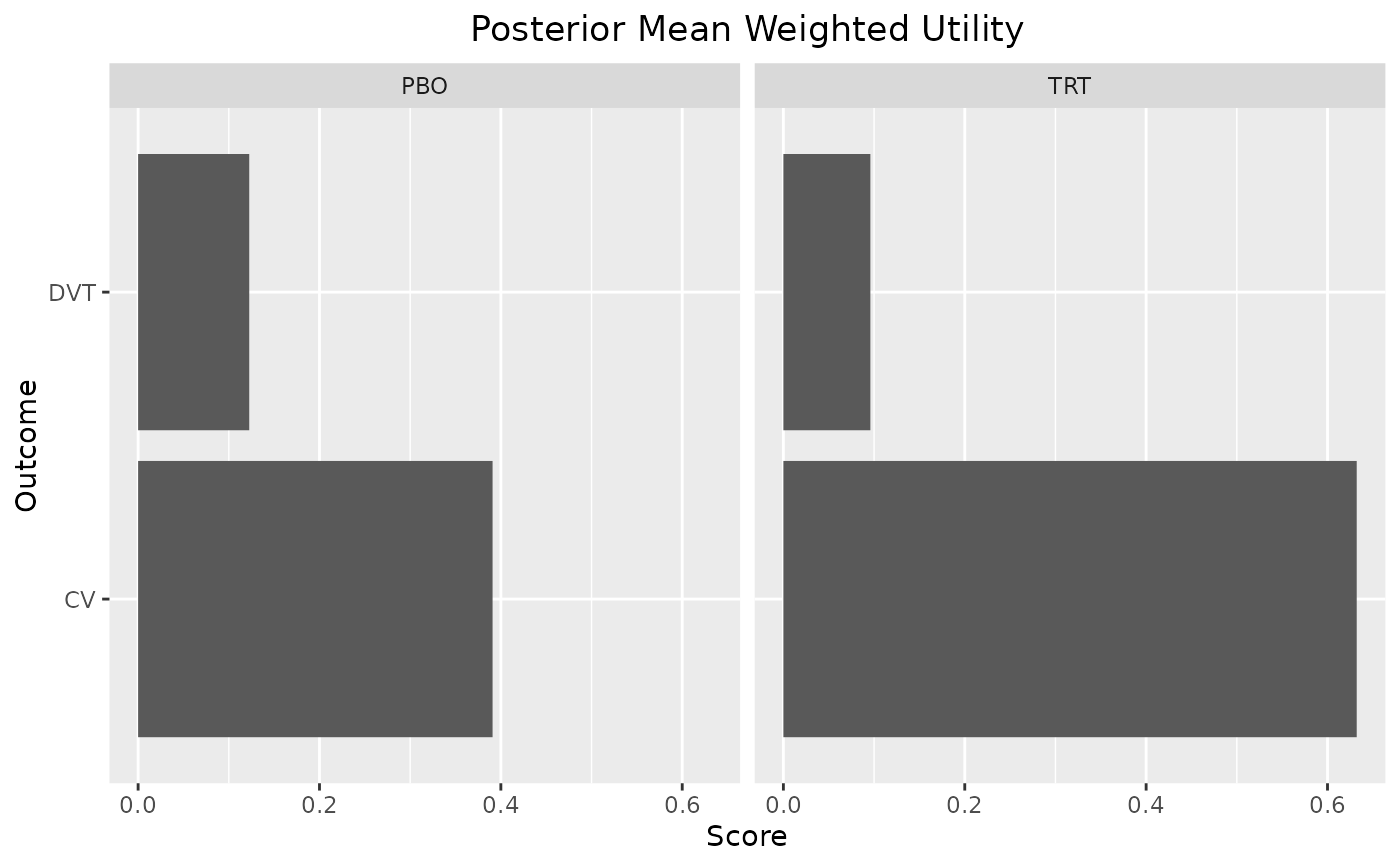
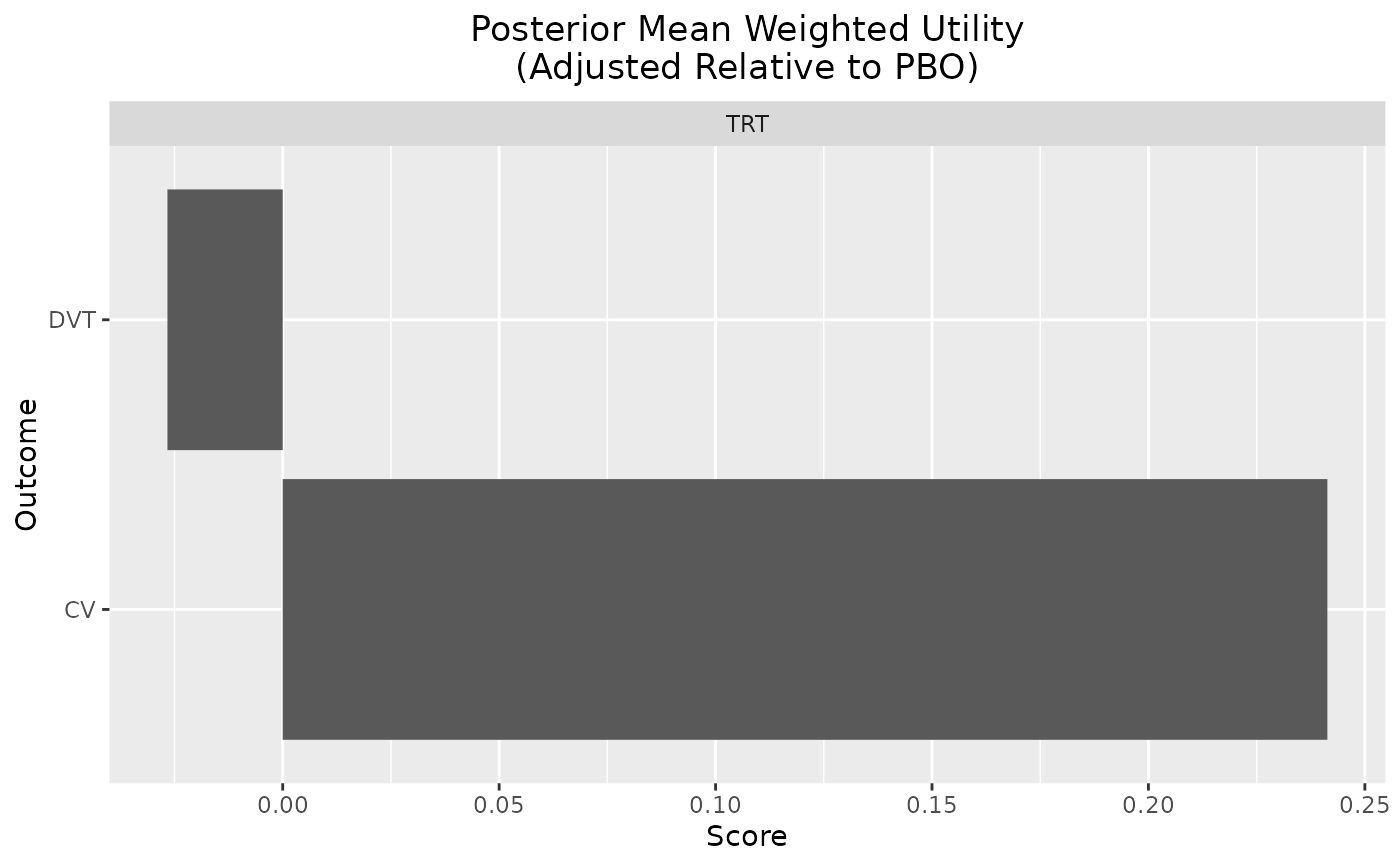 plot_utility(out, stacked = TRUE)
plot_utility(out, stacked = TRUE)